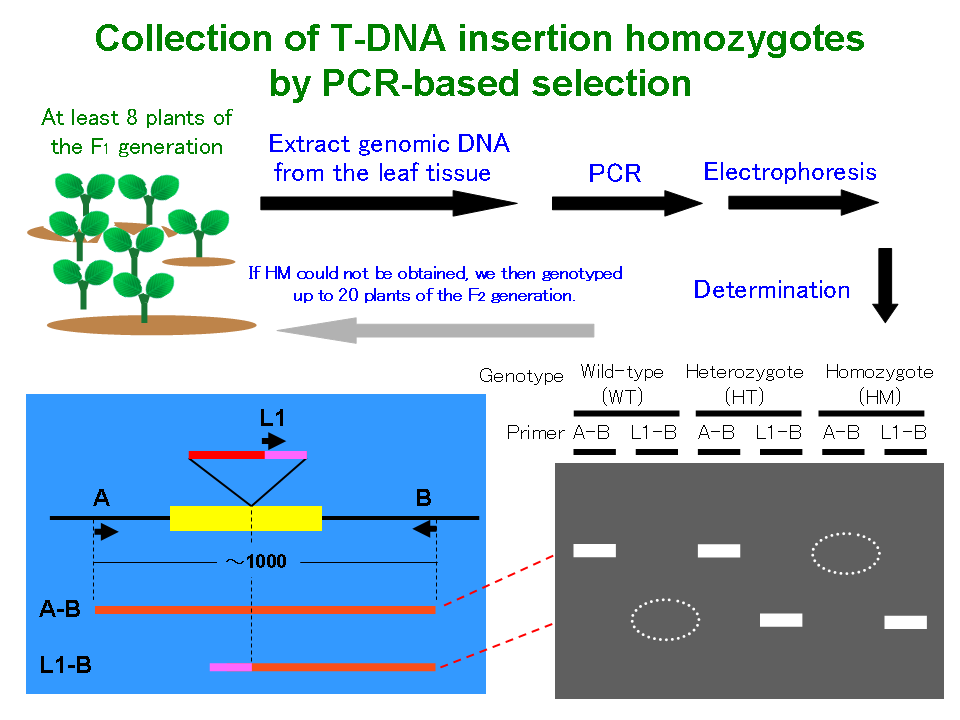
t dna insertion
The T-DNA could be used as a tag to indicate the insertion sites after confirming the existence of exogenous T-DNA in br. The protocol involves the hybridization and capture of sequences containing T-DNA borders using biotinylated oligos.
Plos One Characterization Of Brca2 Deficient Plants Excludes The Role Of Nhej And Ssa In The Meiotic Chromosomal Defect Phenotype
The availability of T-DNA insertion sites is very important for functional genomics research and.

. Authors E Wassim Chehab 1 Se Kim Tatyana Savchenko Daniel Kliebenstein Katayoon Dehesh Janet Braam. Specifically if PCR is performed using one gene-specific primer and one T-DNAspecific primer a PCR product is formed only if a T-DNA element has landed either within or very close to the gene of interest. T-DNA INSERTION SITES IN A.
Were then analyzed by Southern blotting and DNA sequencing. The T -DNA including any sequences inserted between the boarders can be transferred by the. A simple method to identify the sites of transgene insertions using T-DNA-specific primers and high-throughput sequencing that enables identification of multiple insertion sites in the T1 generation of any crop transformed via Agrobacterium.
However mutagenesis allows only partial analysis of genes that function at multiple stages of development genes that are functionally. Sequence-based identification of T-DNA insertion mutations in Arabidopsis. In addition we were able to characterize the status of the T-DNA.
There are several activities which may introduce breaks into plant. Coding sequence CDS exon CDS intron 5UTR. If a large enough population of T-DNAtransformed lines is available one has a very good chance of finding a plant carrying a T-DNA insert within any gene of interest.
Two insertion sites of br were identified and verified as 3156156031610547 bp on Chr2 and 172908 bp1729378 bp on Chr5. Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant Plant Physiol. Transfer DNA T-DNA insertion mutants are often used in forward and reverse genetics to reveal the molecular mechanisms of a particular biological process in plants.
T-DNA - T-DNA line deposited to the ABRC. T-DNA insertion mutagenesis is one of those re- cently developed genetic techniques which can be expected to have a major impact in plant molec- ular biology 17 50 871. When selecting a T-DNA line we prioritize inserts by the following order.
For the example gene At1g01010 the GABI_414G04. For Arabidopsis research community cDNAs please contact individual researchers. The T-DNA may insert in more than one place within the Arabidopsis genome.
This last observation confirms that this transformation approach could be useful to generate dikaryotic. Arabidopsis thaliana deletion expression knockout T-DNA insertion translocation Introduction Transfer-DNA T-DNA insertion is a highly effective mutagen for genome-wide mutagenesis Krysan et al. Under most circumstances seeds for the Salk.
Actin mutants act2-1 and act4-1. Powered by GEBD Please use the backup pageserved by AtTA if the tdnaexpress server is down. Created and developed by Huaming Chen.
Twenty-four of 34 lines appeared to carry T-DNA at one locus while the remaining 10 lines contained unlinked T-DNA insertion Table 1. To generate T-DNA insertion mutants T-DNA must be inserted randomly in the genome through transformation mediated by Agrobacterium tumefaciens. Genetic engineering of crop plants has been successful in transferring traits into elite lines beyond what can be.
The location of the T-DNA insertion relative to gene annotation is the best indicator of whether a gene function will be. Google Scholar Nacry P Camilleri C Courtial B Caboche M Bouchez D. Bicolor strain S238N grown on a medium with nitrate as the only N source.
The results of these DNA sequencing re-actions allowed us to precisely localize the position at which each T-DNA element integrated into the genome. The plant DNA that can be used for insertion of DNA the T-DNA point of. The presence of a T-DNA insertion within any given gene can be easily detected by the proper PCR strategy.
The new tool can return the primers faster and also. The new T-DNA Primer Design Tool is now powered by GenomeExpress Browser Server GEBD. T-DNA insertion mutations are a valuable resource for studies of gene function in Arabidopsis serving as the basis for both forward and reverse genetic strategies and as a source of sequence tags from large collections of mutant lines.
THALIANAAND SATURATION MUTAGENESIS 165. In just a few years plant biology the late starter of molecular genetics has placed itself at the forefront of science in large measure as a result of the more than 20 000 T-DNA transformants that have been. The number of T-DNA insertion loci was analysed by scoring hygromycin-resistant progeny T2 of the primary transgenic plants T1.
Please contact the Arabidopsis Biological Resource Center ABRC to obtain a limited number of seeds for any of the public Salk insertion mutants or SSP UC clones. A schematic representation of T-DNA insertion and tagging system. A further characterization of the phenotype of this mutant with a single T-DNA insertion showed an alteration in hyphal growth rate and number of hyphae compared with the wild-type L.
T-DNA insertion mutagenesis in Arabidopsis has been instrumental in advancing our knowledge of the physiology biochemistry and development of plants. T-DNA Express - T-DNA verification primer design SIGNAL. The purpose of this method is to localize T-DNA insertion sites in transgenic plants.
View or for DNA repair the plant point of view. T-DNA_Insertion_Identify Identifying T-DNA insertion sites by whole-genome resequencing 利用重测序技术鉴定 T-DNA 插入位点. This indicates that transgenic plants contain an average of 14 loci of T-DNA inserts.
T-DNA - T-DNA line not yet deposited. This procedure was tested with DNA from Oryza sativa L. Unlike conventional re- views this brief outlook intends to give a practi- cal insight into current problems and future applications of T-DNA tagging by underlining the.
17 of the time compared to 8 in insertion before the start codon. T-DNA insertion can also cause deletion and translocation. Due to the nature of the T-DNA integration event and the limitations of the PCR border recovery protocol PCR products are sequenced directly without separating the products of each reaction.
Epub 2011 Apr 12. Affiliation 1 Rice University. Cause there is very little intergenic material the insertion of a piece of T-DNA on the order of 5 to 25 kb in length generally produces a dramatic disruption of gene function.
Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis.

Molecular Characterization Of Pht4 2 T Dna Insertion Mutants A Scheme Download Scientific Diagram

Identification Of T Dna Insertion Lines For Two Atmsr Genes A Gene Download Scientific Diagram

T Dna Insertion Mutation In The Gcr1 Gene In Arabidopsis Thaliana And Download Scientific Diagram

T Dna Insertion And Expression In Lig6 Mutant A Position Of The Download Scientific Diagram

Agrobacterium T Dna Integration Molecules And Models Trends In Genetics

Fig 7 Functional Analysis Of The Arabidopsis Centromere By T Dna Insertion Induced Centromere Breakage Pnas
Chloroplast Function Database Iii About
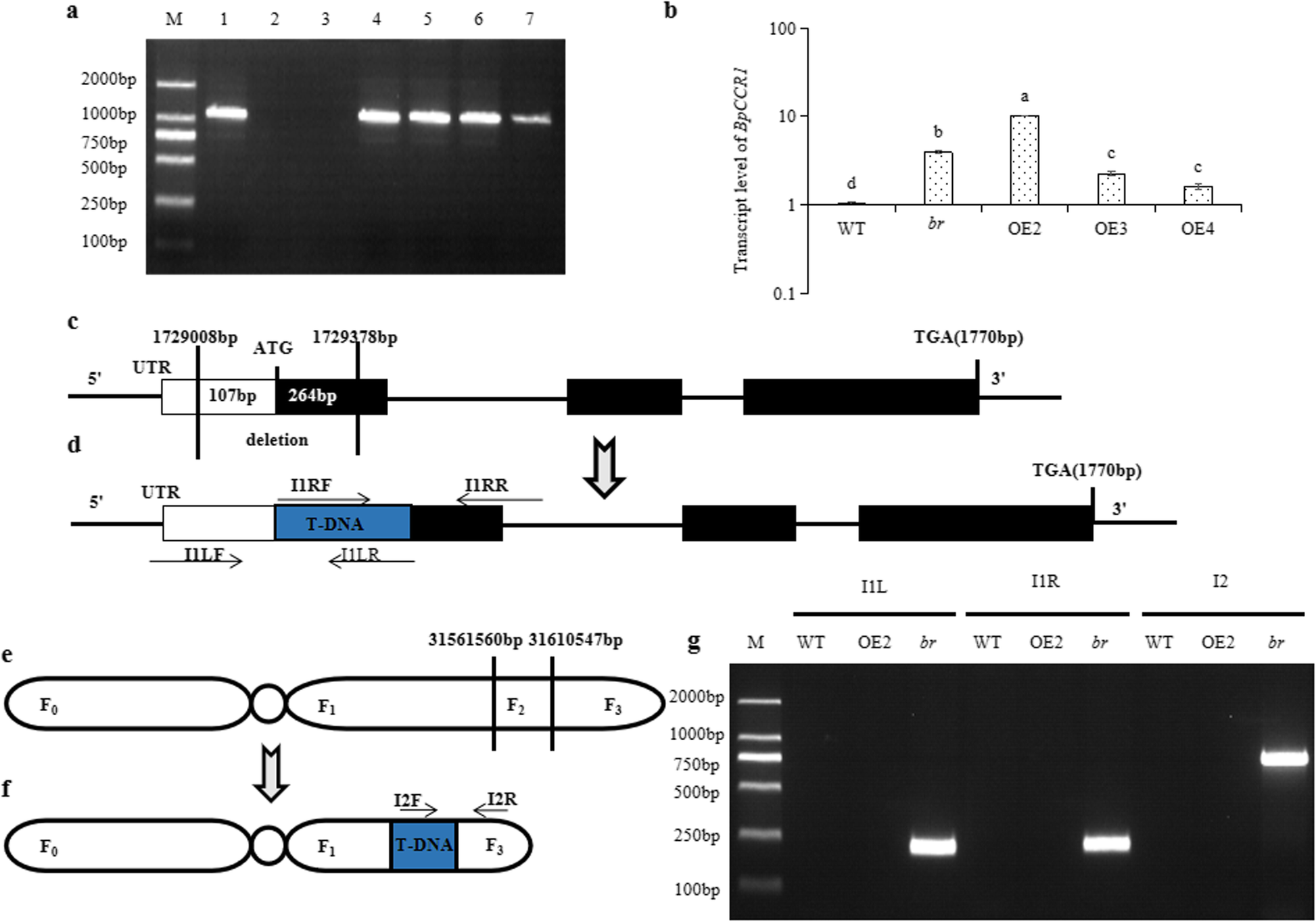
Characterization And T Dna Insertion Sites Identification Of A Multiple Branches Mutant Br In Betula Platyphylla Betula Pendula Bmc Plant Biology Full Text
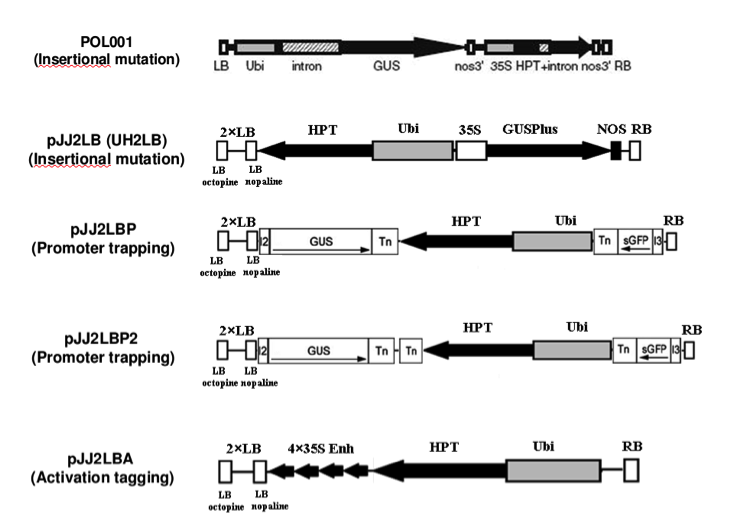
Brachypodium T Dna Collection Doe Joint Genome Institute

Molecular Analysis And The Effect Of The T Dna Insertion In Mre11 Mutant Lines

Csp41a T Dna Insertion Lines Used In This Study A Scheme Of The A Download Scientific Diagram

Pcr Genotyping For Analysis Of T Dna Insertion Mutant Plants At Dna Download Scientific Diagram
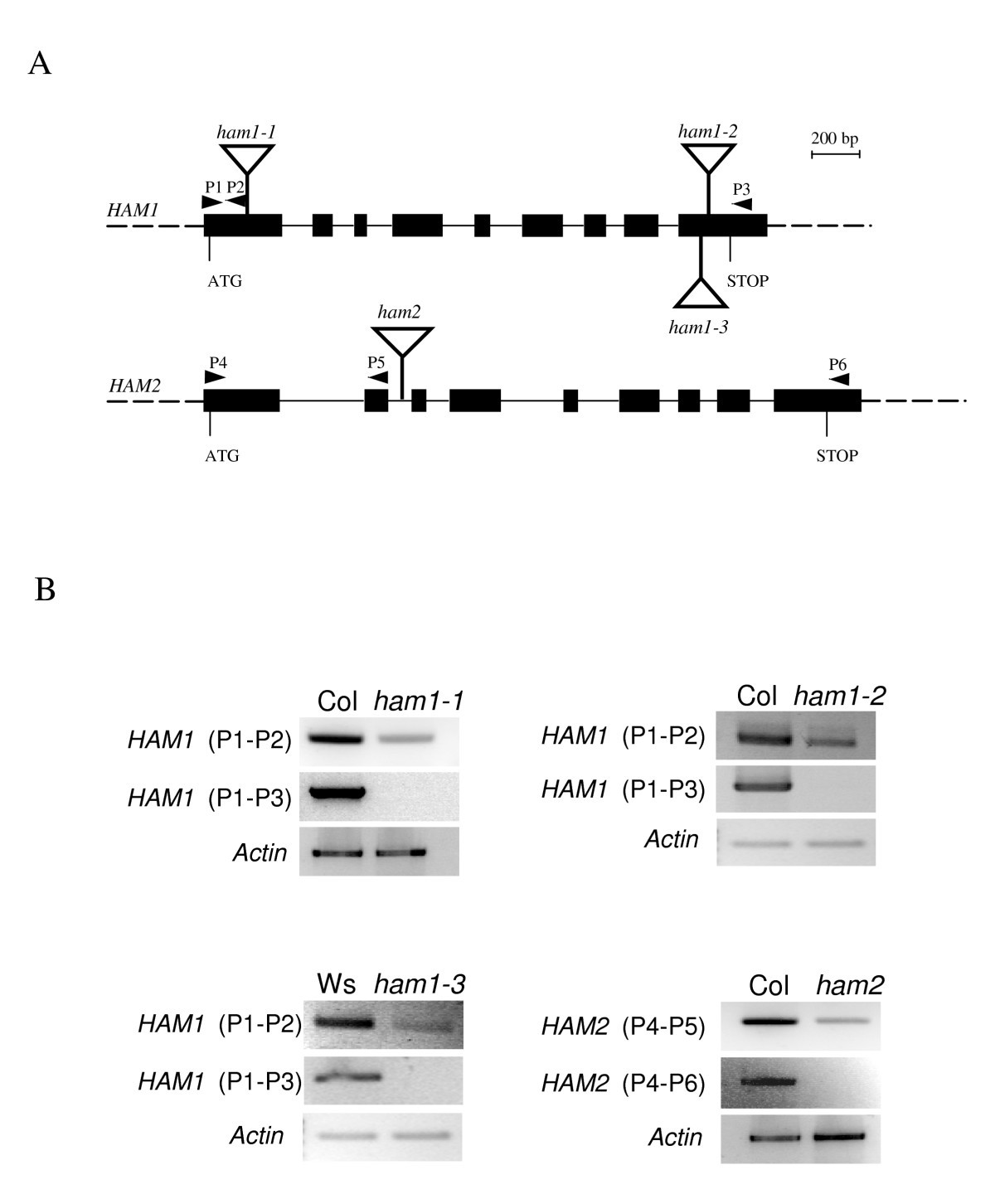
The Myst Histone Acetyltransferases Are Essential For Gametophyte Development In Arabidopsis Bmc Plant Biology Full Text

Fig S1 Overview Of T Dna Insertion Lines Isolated And Used In This Download Scientific Diagram

Figure S3 Characterisation Of Arabidopsis T Dna Insertion Mutants A Download Scientific Diagram
Plos Computational Biology Eat Rice A Predictive Model For Flanking Gene Expression Of T Dna Insertion Activation Tagged Rice Mutants By Machine Learning Approaches

Characterization Of T Dna Insertion Mutants And Rnai Silenced Plants Of Arabidopsis Thaliana Uv Damaged Dna Binding Protein 2 Atuv Ddb2 Semantic Scholar
Comments
Post a Comment